GPCRM Models
- File: 5HT1A_HUMAN_inact.zip
- File: 5HT1B_HUMAN_inact.zip
- File: 5HT1D_HUMAN_inact.zip
- File: 5HT1E_HUMAN_inact.zip
- File: 5HT1F_HUMAN_inact.zip
- File: 5HT2A_HUMAN_inact.zip
- File: 5HT2B_HUMAN_inact.zip
- File: 5HT2C_HUMAN_inact.zip
- File: 5HT4R_HUMAN_inact.zip
- File: 5HT6R_HUMAN_inact.zip
- File: AA1R_HUMAN_inact.zip
- File: AA2AR_HUMAN_inact.zip
- File: AA2BR_HUMAN_inact.zip
- File: AA3R_HUMAN_inact.zip
- File: ACM1_HUMAN_inact.zip
- File: ACM2_HUMAN_inact.zip
- File: ADA1A_HUMAN_inact.zip
- File: ADA1B_HUMAN_inact.zip
- File: ADA1D_HUMAN_inact.zip
- File: ADA2A_HUMAN_inact.zip
- File: ADA2C_HUMAN_inact.zip
- File: ADRB2_HUMAN_inact.zip
- File: ADRB3_HUMAN_inact.zip
- File: AGTR1_HUMAN_inact.zip
- File: C3AR_HUMAN_inact.zip
- File: CCBP2_HUMAN_inact.zip
- File: CCR1_HUMAN_inact.zip
- File: CCR2_HUMAN_inact.zip
- File: CCR4_HUMAN_inact.zip
- File: CCR5_HUMAN_inact.zip
- File: CCR6_HUMAN_inact.zip
- File: CCR7_HUMAN_inact.zip
- File: CCR9_HUMAN_inact.zip
- File: CCRL1_HUMAN_inact.zip
- File: CLTR1_HUMAN_inact.zip
- File: CML1_HUMAN_inact.zip
- File: CNR1_HUMAN_inact.zip
- File: CXCR1_HUMAN_inact.zip
- File: CXCR2_HUMAN_inact.zip
- File: CXCR3_HUMAN_inact.zip
- File: CXCR4_HUMAN_inact.zip
- File: CXCR5_HUMAN_inact.zip
- File: CXCR6_HUMAN_inact.zip
- File: DRD1_HUMAN_inact.zip
- File: DRD2_HUMAN_inact.zip
- File: DRD3_HUMAN_inact.zip
- File: DRD4_HUMAN_inact.zip
- File: DRD5_HUMAN_inact.zip
- File: EDNRA_HUMAN_inact.zip
- File: FPR1_HUMAN_inact.zip
- File: FSHR_HUMAN_inact.zip
- File: GALR1_HUMAN_inact.zip
- File: GHSR_HUMAN_inact.zip
- File: GNRHR_HUMAN_inact.zip
- File: GPR44_HUMAN_inact.zip
- File: GPR4_HUMAN_inact.zip
- File: GPR56_HUMAN_inact.zip
- File: GPR85_HUMAN_inact.zip
- File: HCAR3_HUMAN_inact.zip
- File: HRH3_HUMAN_inact.zip
- File: LPAR1_HUMAN_inact.zip
- File: LPAR3_HUMAN_inact.zip
- File: LPAR6_HUMAN_inact.zip
- File: LSHR_HUMAN_inact.zip
- File: LT4R1_HUMAN_inact.zip
- File: MAS_HUMAN_inact.zip
- File: MC4R_HUMAN_inact.zip
- File: MCHR1_HUMAN_inact.zip
- File: MRGX1_HUMAN_inact.zip
- File: MTR1A_HUMAN_inact.zip
- File: MTR1L_HUMAN_inact.zip
- File: NMBR_HUMAN_inact.zip
- File: NPBW1_HUMAN_inact.zip
- File: NPFF2_HUMAN_inact.zip
- File: NPY1R_HUMAN_inact.zip
- File: NTR1_HUMAN_inact.zip
- File: OPN4_HUMAN_inact.zip
- File: OPRD_HUMAN_inact.zip
- File: OPRK_HUMAN_inact.zip
- File: OPRM_HUMAN_inact.zip
- File: OPRX_HUMAN_inact.zip
- File: OPSB_HUMAN_inact.zip
- File: OPSB_HUMAN_inact_alternative_alignment.zip
- File: OPSD_HUMAN_inact.zip
- File: OPSD_HUMAN_inact_alternative_alignment.zip
- File: OPSG_HUMAN_inact.zip
- File: OPSG_HUMAN_inact_alternative_alignment.zip
- File: OPSR_HUMAN_inact.zip
- File: OR1A1_HUMAN_alternative_alignment_inact.zip
- File: OR1A1_HUMAN_inact.zip
- File: OR1D2_HUMAN_inact.zip
- File: OR1G1_HUMAN_inact.zip
- File: OR3A1_HUMAN_alternative_alignment_inact.zip
- File: OR3A1_HUMAN_inact.zip
- File: OX1R_HUMAN_inact.zip
- File: P2RY1_HUMAN_alternative_alignment_inact.zip
- File: P2RY1_HUMAN_inact.zip
- File: P2Y12_HUMAN_inact.zip
- File: P2Y12_HUMAN_inact_alternative_alignment.zip
- File: PAR1_HUMAN_inact.zip
- File: PAR1_HUMAN_inact_alternative_alignment.zip
- File: PD2R_HUMAN_alternative_alignment_inact.zip
- File: PD2R_HUMAN_inact.zip
- File: Q4VBL8_HUMAN_inact.zip
- File: SSR1_HUMAN_inact.zip
- File: SSR4_HUMAN_inact.zip
- File: SSR4_HUMAN_inact_alternative_alignment.zip
- File: TA2R_HUMAN_inact.zip
- File: TA2R_HUMAN_inact_alternative_alignment.zip
- File: TAAR1_HUMAN_inact.zip
- File: TAAR8_HUMAN_inact.zip
- File: V1AR_HUMAN_inact.zip
- File: V2R_HUMAN_inact.zip
- File: XCR1_HUMAN_alternative_alignment_inact.zip
- File: XCR1_HUMAN_inact.zip
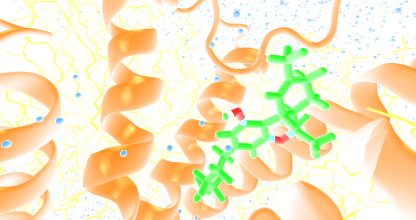
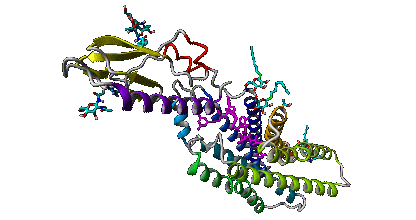
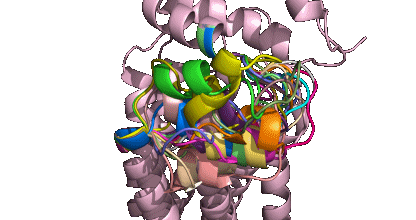
To generate the above theoretical models of GPCRs from the rhodopsin-like subfamily we used GPCRM with 'Auto mode' and 'short loop modeling' option.
We strongly advice you to try GPCRM server for more accurate loop modeling.
1st and 2nd target datasets refer to our PLOS 2013 paper.