List of templates
Set of active templates structures:
| # | PDB CODE (CHAIN) |
UNIPROT CODE |
ORGANISM | RECEPTOR SUBFAMILY-TYPE |
RESOLUTION [Å] |
# AMINO ACIDS (PDB/UNIPROT) |
FUSION PROTEIN | FIRST AUTHOR | JOURNAL | YEAR | LIGAND CODE |
LIGAND ROLE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2Y02 (B) | P07700 | turkey | adrenergic-beta1 | 2.60 | 298/483 | none | A. Warne | NATURE | 2011 | WHJ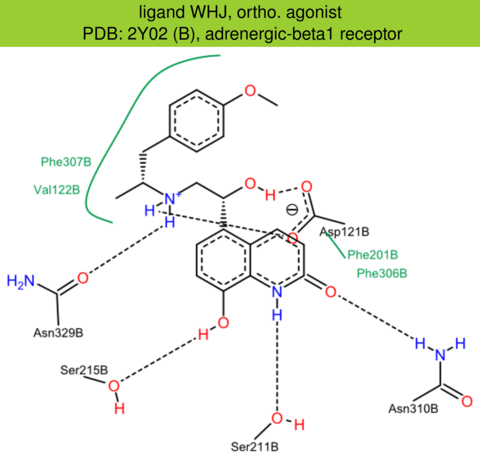 |
ortho. agonist |
| 2 | 2YDV (A) | P29274 | human | adenosine-A2A | 2.60 | 307/412 | none | G. Lebon | NATURE | 2011 | NEC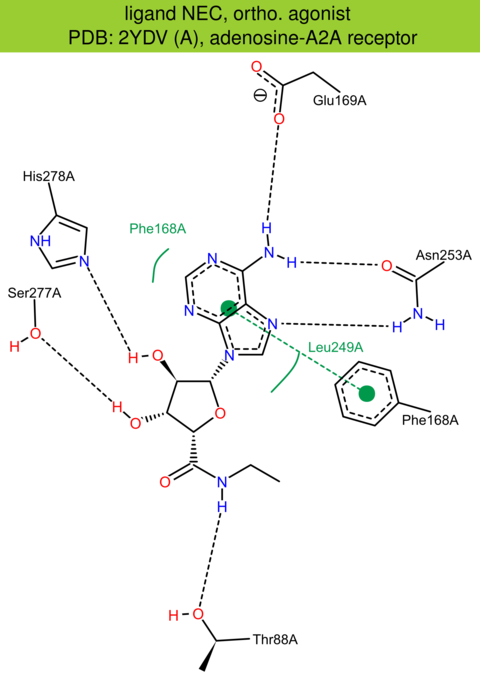 |
ortho. agonist |
| 3 | 3PQR (A) | P02699 | bovine | rhodopsin | 2.85 | 326/348 | none | H.W. Choe | NATURE | 2011 | RET |
ortho. agonist |
| 4 | 3QAK (A) | P29274 | human | adenosine-A2A | 2.71 | 284/412 | lysozyme | F. Xu | SCIENCE | 2011 | UKA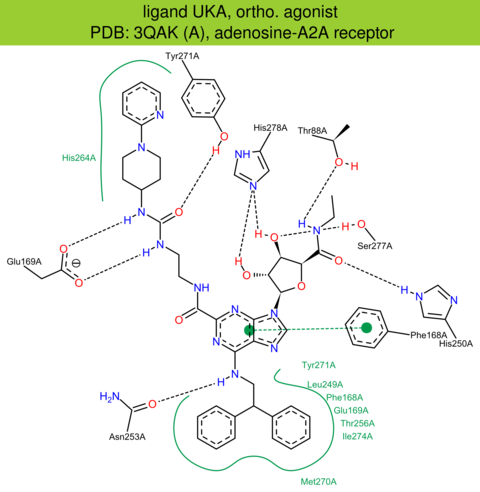 |
ortho. agonist |
| 5 | 3SN6 (R) | P04896 | bovine | adrenergic-beta2 | 3.20 | 284/394 | lysozyme | S.G. Rasmussen | NATURE | 2011 | P0G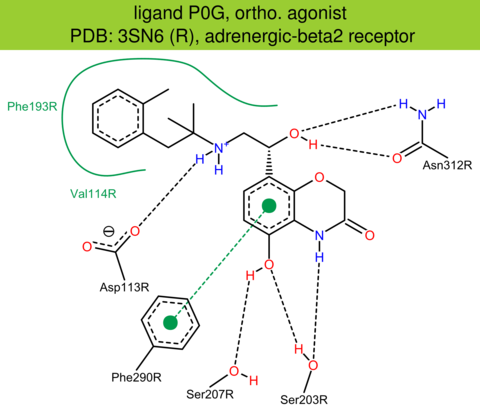 |
ortho. agonist |
| 6 | 4GRV (A) | P20789 | rat | neurotensin-R1 | 2.80 | 293/424 | lysozyme | J.F. White | NATURE | 2012 | PEPTIDE |
ortho. agonist |
| 7 | 4IAQ (A) | P28222 | human | serotonin-1B | 2.80 | 273/390 | cytochrome | C. Wang | SCIENCE | 2013 | 2GM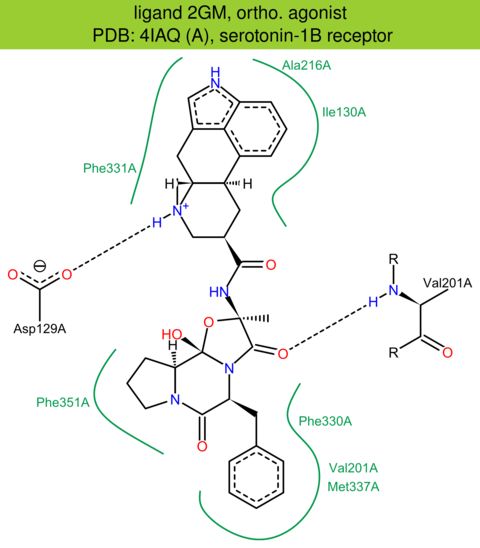 |
ortho. agonist |
| 8 | 4IAR (A) | P28222 | human | serotonin-1B | 2.70 | 273/390 | cytochrome | C. Wang | SCIENCE | 2013 | ERM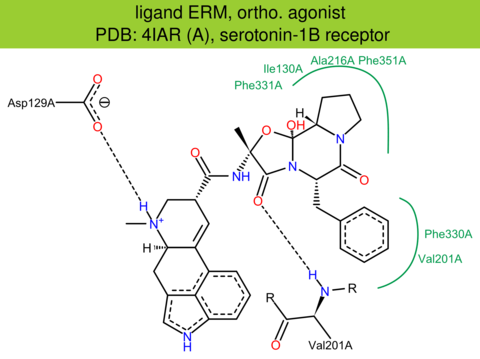 |
ortho. agonist |
| 9 | 4IB4 (A) | P41595 | human | serotonin-2B | 2.70 | 285/481 | cytochrome | D. Wacker | SCIENCE | 2013 | ERM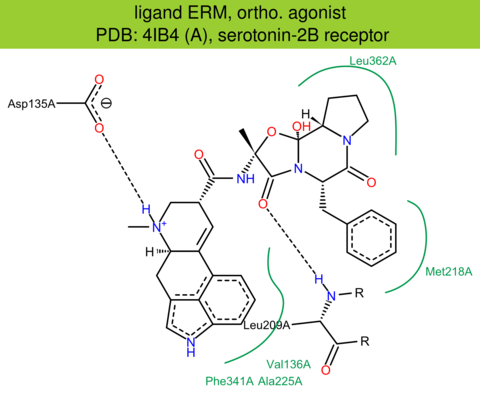 |
ortho. agonist |
| 10 | 4NC3 (A) | P41595 | human | serotonin-2B | 2.80 | 287/481 | cytochrome | W. Liu | SCIENCE | 2013 | ERM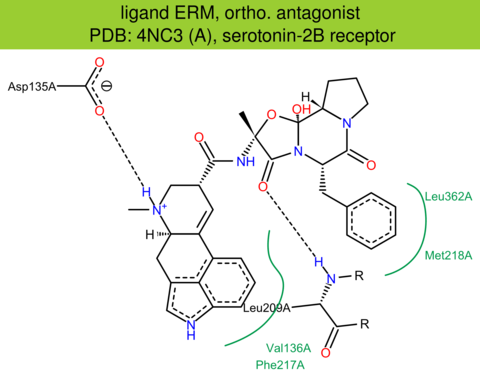 |
ortho. agonist |
| 11 | 4PHU (A) | O14842 | human | fatty-acid-R1 | 2.33 | 272/300 | lysozyme | A. Srivastava | NATURE | 2014 | 2YB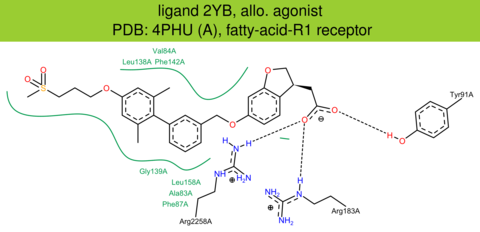 |
allo. agonist |
| 12 | 4PXZ (A) | Q9H244 | human | purinergic-P2Y12 | 2.50 | 286/342 | cytochrome | J. Zhang | NATURE | 2014 | 6AD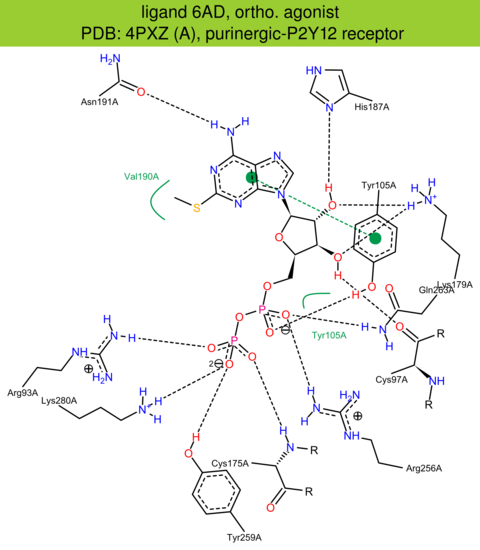 |
ortho. agonist |
| 13 | 4PY0 (A) | Q9H244 | human | purinergic-P2Y12 | 3.10 | 283/342 | cytochrome | J. Zhang | NATURE | 2014 | 6AT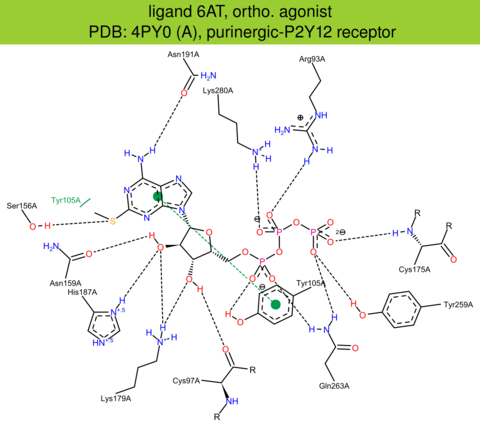 |
ortho. agonist |
| 14 | 4QIN (A) | Q99835 | human | smoothened | 2.60 | 341/787 | cytochrome | C. Wang | NAT. COMMUN. | 2014 | SG8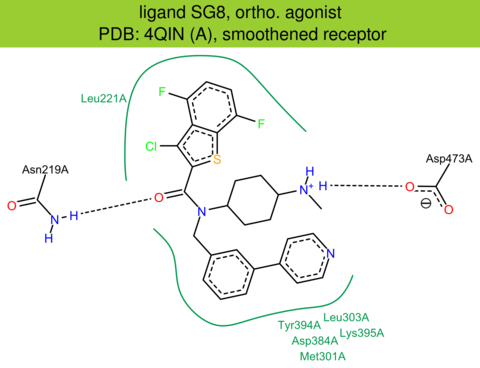 |
ortho. agonist |
| 15 | 4UHR (A) | P29274 | human | adenosine-A2A | 2.60 | 308/412 | none | G. Lebon | MOL. PHARMACOL. | 2015 | NGI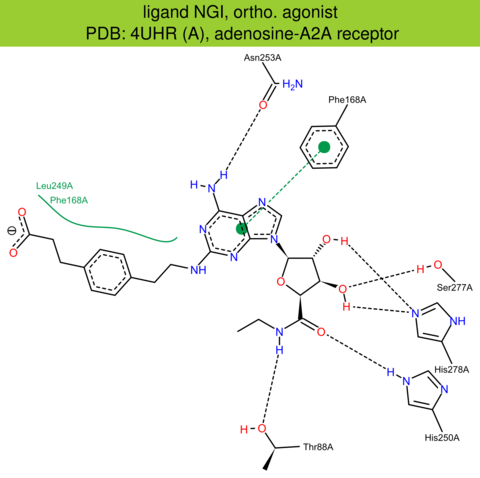 |
ortho. agonist |
| 16 | 4XT1 (A) | P69332 | human | chemokine-CX3CL1 | 2.89 | 291/354 | none | J.S. Burg | SCIENCE | 2015 | protein |
ortho. agonist |
| 17 | 4XT3 (A) | P69332 | human | chemokine-CX3CL1 | 3.80 | 288/354 | none | J.S. Burg | SCIENCE | 2015 | protein |
ortho. agonist |
| 18 | 5C1M (A) | P42866 | mouse | opioid-mu | 2.10 | 295/398 | none | W. Huang | NATURE | 2015 | 4VO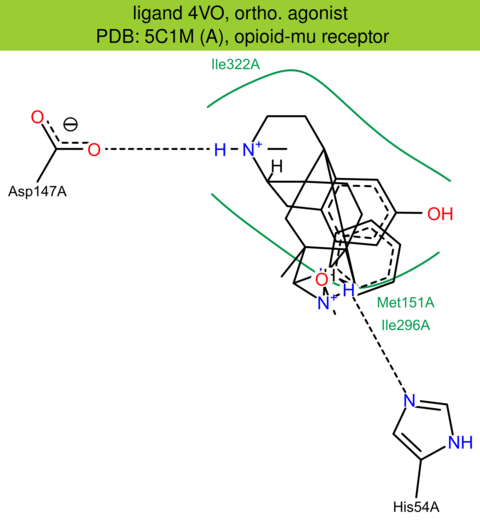 |
ortho. agonist |
| 19 | 5G53 (A) | P29274 | human | adenosine-A2A | 3.40 | 279/412 | none | B. Carpenter | NATURE | 2016 | NEC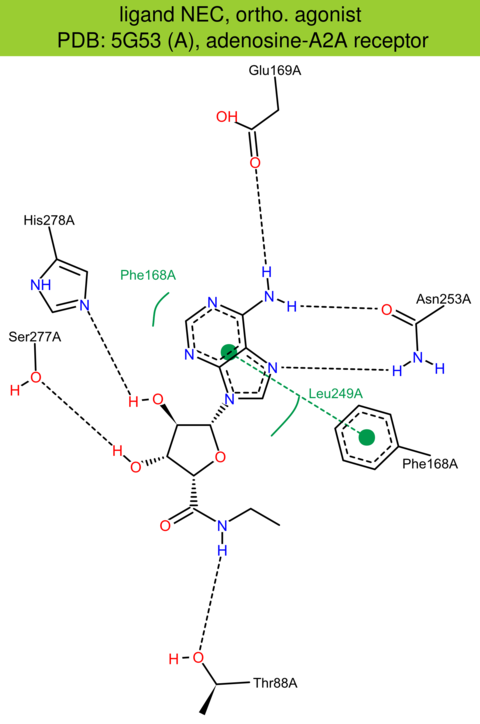 |
ortho. agonist |
| 20 | 5GLH (A) | P24530 | human | endothelin-B | 2.80 | 292/442 | none | W. Shihoya | NATURE | 2016 | peptide |
ortho. agonist |
| 21 | 5NX2 (A) | P43220 | human | 5NX2_A:* | 3.70 | 389/463 | none | A. Jazayeri | NATURE | 2017 | none | none |
| 22 | 5TVN (A) | P41595 | human | serotonin-2B | 2.90 | 293/481 | cytochrome | D. Wacker | CELL | 2017 | 7LD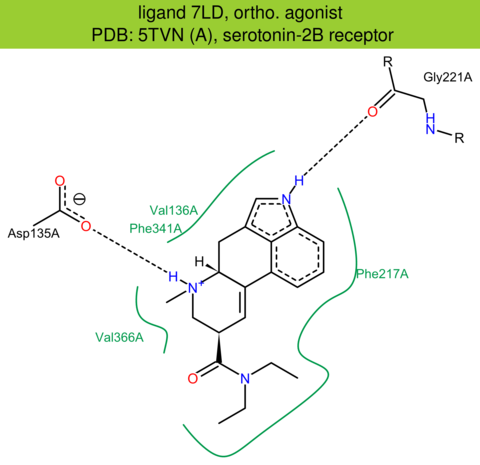 |
ortho. agonist |
| 23 | 5UNG (A) | P50052 | human | angiotensin-AT2R | 2.80 | 300/363 | cytochrome | H. Zhang | NATURE | 2017 | 8ES |
ortho. ligand |
| 24 | 5UZ7 (A) | P63092 | human | calcitonin | 4.10 | 266/394 | none | Y.L. Liang | NATURE | 2017 | none | none |
| 25 | 5VAI (R) | G1SGD4 | rabbit | glucagon-like-R1 | 4.10 | 387/462 | none | Y. Zhang | NATURE | 2017 | none | none |
| 26 | 5VBL (A) | P35414 | human | apelin-R | 2.60 | 299/380 | ruberodoxin | Y. Ma | STRUCTURE | 2017 | none | none |
| 27 | 5XR8 (A) | P21554 | human | cannabinoid-CB1 | 2.95 | 285/472 | flavodoxin | T. Hua | NATURE | 2017 | 8D0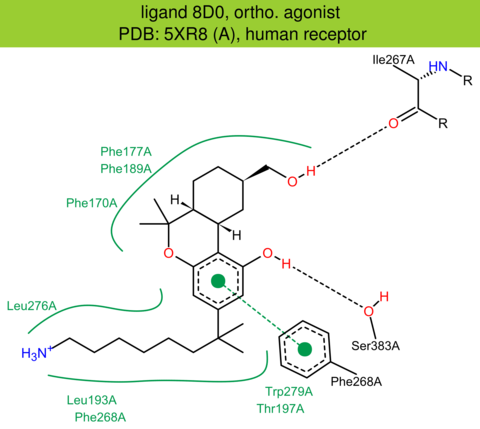 |
ortho. agonist |
| 28 | 5XRA (A) | P21554 | human | cannabinoid-CB1 | 2.80 | 282/472 | flavodoxin | T. Hua | NATURE | 2017 | 8D3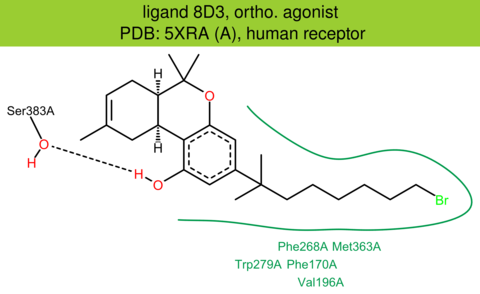 |
ortho. agonist |
| 29 | 5YQZ (R) | P47871 | human | glucagon | 3.00 | 398/477 | lysozyme | H. Zhang | NATURE | 2018 | none | none |
| 30 | 6B73 (A) | P41145 | human | opioid-kappa | 3.10 | 278/380 | cytochrome | T. Che | CELL | 2018 | CVV |
ortho. agonist |
| 31 | 6BQG (A) | P28335 | human | serotonin-2C | 3.00 | 286/458 | cytochrome | Y. Peng | CELL | 2018 | ERM |
ortho. agonist |
Set of inactive templates structures:
| # | PDB CODE (CHAIN) |
UNIPROT CODE |
ORGANISM | RECEPTOR SUBFAMILY-TYPE |
RESOLUTION [Å] |
# AMINO ACIDS (PDB/UNIPROT) |
FUSION PROTEIN | FIRST AUTHOR | JOURNAL | YEAR | LIGAND CODE |
LIGAND ROLE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1GZM (A) | P02699 | bovine | rhodopsin | 2.65 | 329/348 | none | J. Li | J. MOL. BIOL. | 2004 | RET |
ortho. antagonist |
| 2 | 1U19 (A) | P02699 | cattle | rhodopsin | 2.20 | 348/348 | none | T. Okada | J. MOL. BIOL. | 2004 | RET |
ortho. antagonist |
| 3 | 2RH1 (A) | P07550 | human | adrenergic-beta2 | 2.40 | 282/413 | lysozyme | V. Cherezov | SCIENCE | 2007 | CAU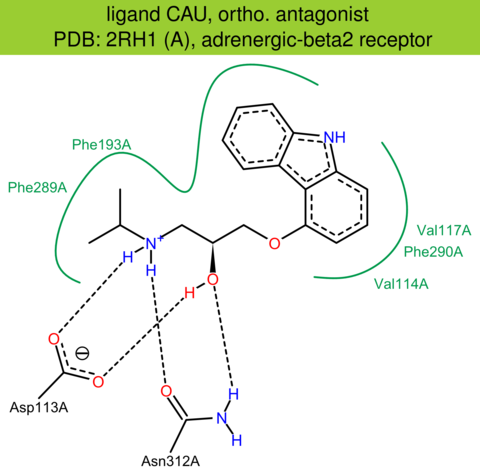 |
ortho. antagonist |
| 4 | 2VT4 (A) | P07700 | turkey | adrenergic-beta1 | 2.7 | 276/483 | lysozyme | A. Warne | NATURE | 2008 | P32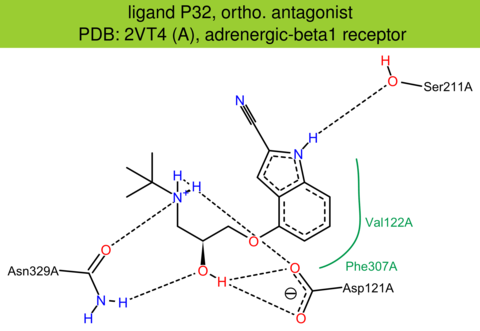 |
ortho. antagonist |
| 5 | 3EML (A) | P29274 | human | adenosine-A2A | 2.60 | 288/412 | lysozyme | V.P. Jaakola | SCIENCE | 2008 | ZMA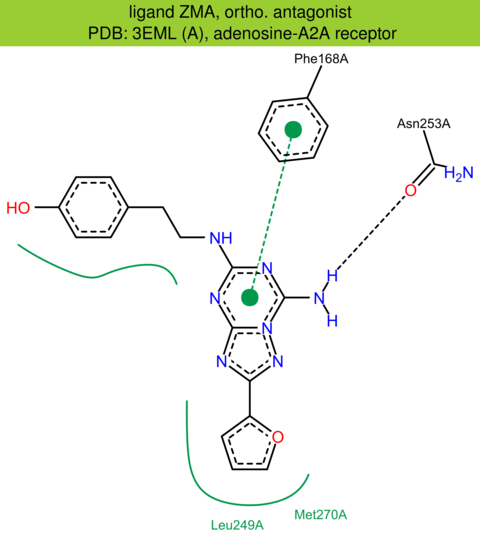 |
ortho. antagonist |
| 6 | 3PBL (A) | P35462 | human | dopamine-D3 | 2.89 | 272/400 | lysozyme | E.Y. Chien | SCIENCE | 2010 | ETQ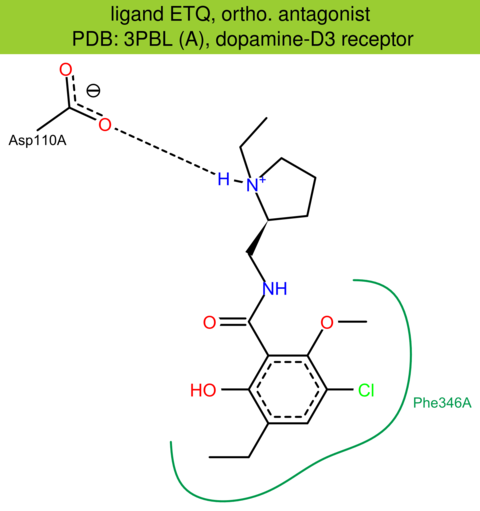 |
ortho. antagonist |
| 7 | 3RZE (A) | P35367 | human | histamine-H1 | 3.10 | 268/487 | lysozyme | T. Shimamura | NATURE | 2011 | 5EH/D7V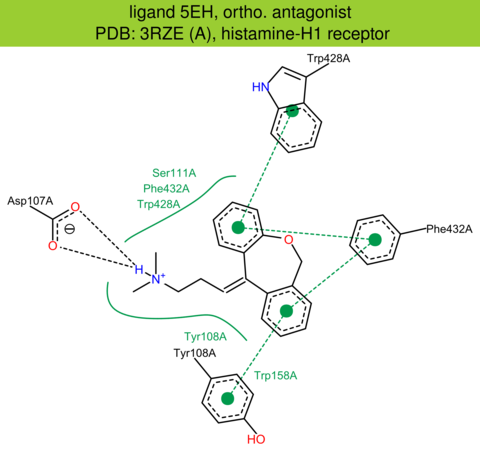 |
ortho. antagonist |
| 8 | 3UON (A) | P08172 | human | muscarinic-M2 | 3.00 | 278/466 | lysozyme | K. Haga | NATURE | 2012 | QNB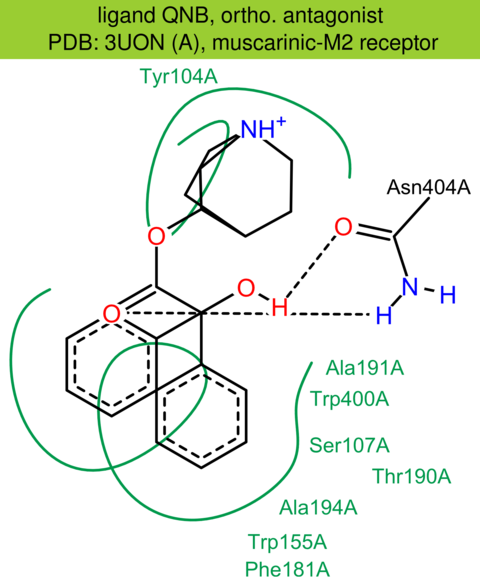 |
ortho. antagonist |
| 9 | 3V2Y (A) | P21453 | human | sphingosine-R1 | 2.80 | 291/382 | lysozyme | M.A. Hanson | SCIENCE | 2012 | ML5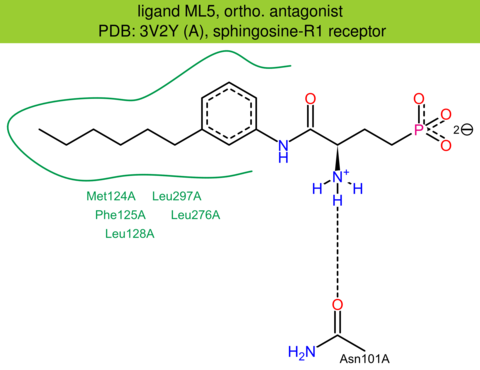 |
ortho. antagonist |
| 10 | 3VW7 (A) | P25116 | human | protease-activated-R1 | 2.20 | 282/425 | lysozyme | C. Zhang | NATURE | 2012 | VPX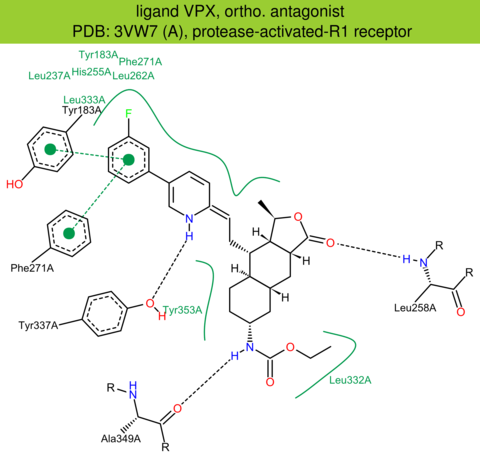 |
ortho. antagonist |
| 11 | 4DAJ (A) | P08483 | rat | muscarinic-M3 | 3.40 | 268/589 | lysozyme | A.C. Kruse | NATURE | 2012 | 0HK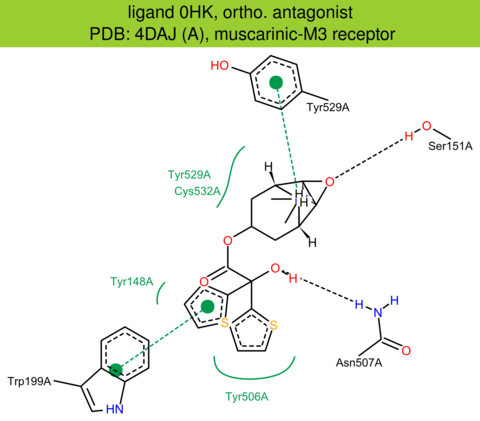 |
ortho. antagonist |
| 12 | 4DJH (A) | P41145 | human | opioid-kappa | 2.90 | 287/380 | lysozyme | H. Wu | NATURE | 2012 | JDC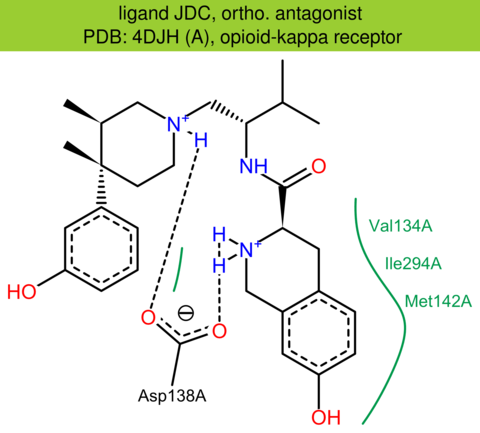 |
ortho. antagonist |
| 13 | 4DKL (A) | P42866 | mouse | opioid-mu | 2.80 | 282/398 | lysozyme | A. Manglik | NATURE | 2012 | BF0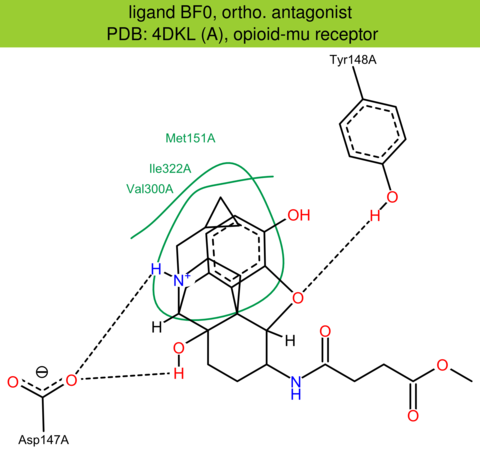 |
ortho. antagonist |
| 14 | 4EA3 (A) | P41146 | human | opioid-nociceptin | 3.01 | 278/370 | cytochrome | A.A. Thompson | NATURE | 2012 | 0NN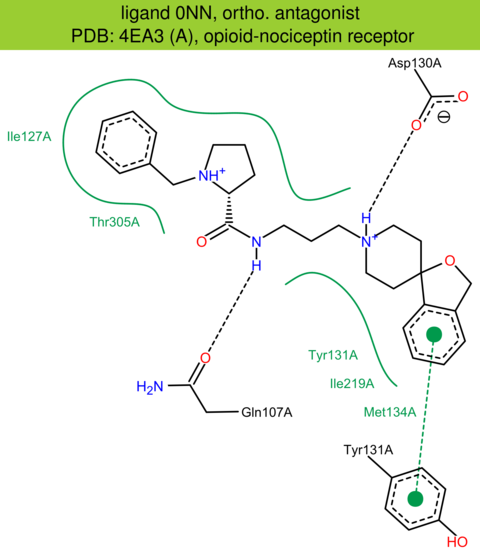 |
ortho. antagonist |
| 15 | 4EIY (A) | P29274 | human | adenosine-A2A | 1.80 | 296/412 | cytochrome | W. Liu | SCIENCE | 2012 | ZMA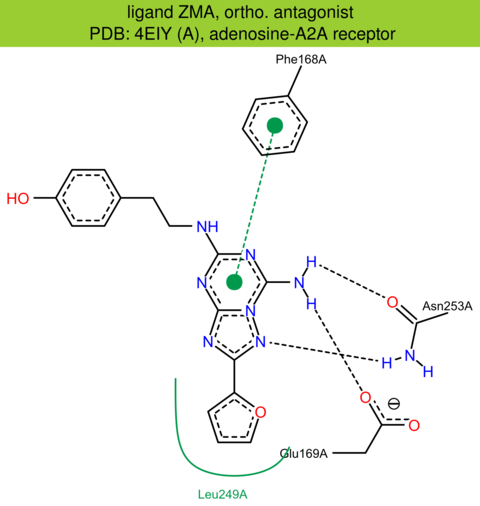 |
ortho. antagonist |
| 16 | 4EJ4 (A) | P32300 | mouse | opioid-delta | 3.40 | 282/372 | lysozyme | S. Granier | NATURE | 2012 | EJ4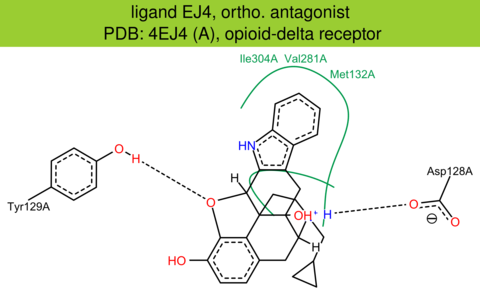 |
ortho. antagonist |
| 17 | 4JKV (A) | Q99835 | human | smoothened | 2.45 | 346/787 | cytochrome | C. Wang | NATURE | 2013 | 1KS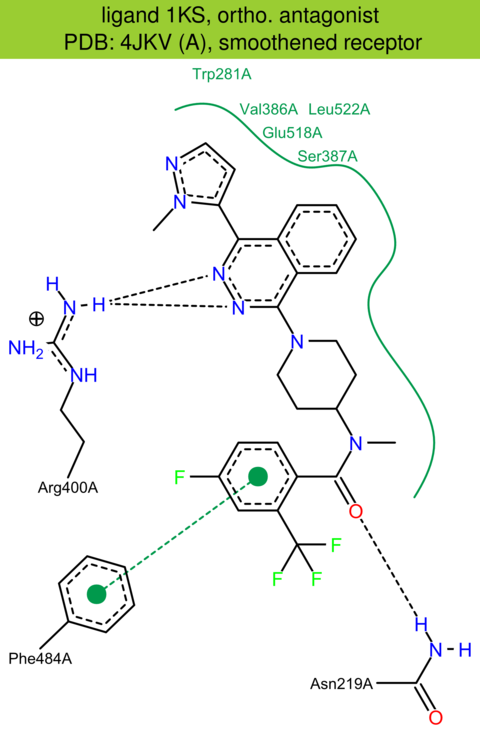 |
ortho. antagonist |
| 18 | 4K5Y (A) | P34998 | human | corticotropin-R1 | 2.98 | 247/444 | lysozyme | K. Hollenstein | NATURE | 2013 | 1Q5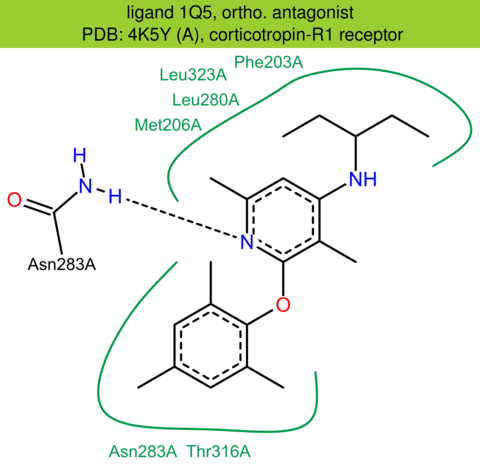 |
ortho. antagonist |
| 19 | 4L6R (A) | P47871 | human | glucagon | 3.30 | 292/477 | cytochrome | F.Y. Siu | NATURE | 2013 | none | none |
| 20 | 4MBS (A) | P51681 | human | chemokine-CCR5 | 2.71 | 292/352 | ruberodoxin | Q. Tan | SCIENCE | 2013 | MRV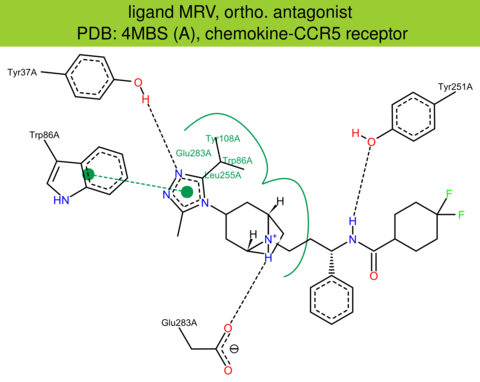 |
ortho. antagonist |
| 21 | 4N4W (A) | Q0SXH8 | human | smoothened | 2.80 | 348/787 | cytochrome | C. Wang | NAT. COMMUN. | 2014 | SNT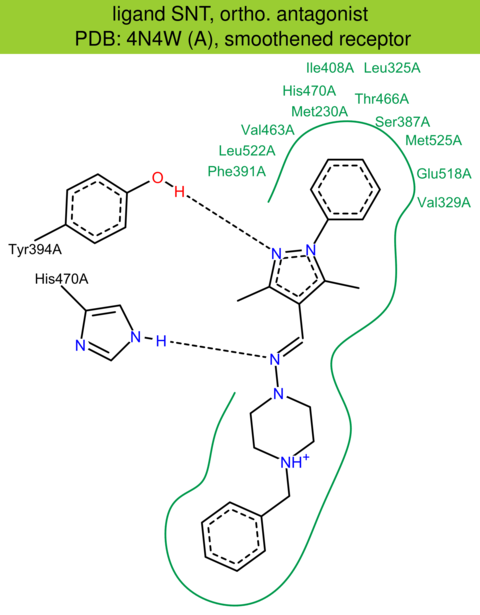 |
ortho. antagonist |
| 22 | 4N6H (A) | P41143 | human | opioid-delta | 1.80 | 303/372 | cytochrome | G. Fenalti | NATURE | 2014 | EJ4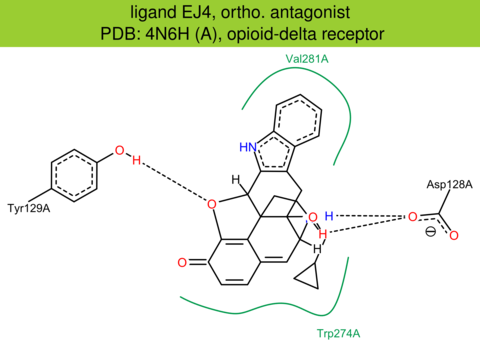 |
ortho. antagonist |
| 23 | 4NTJ (A) | Q9H244 | human | purinergic-P2Y12 | 2.62 | 267/342 | cytochrome | K. Zhang | NATURE | 2014 | AZJ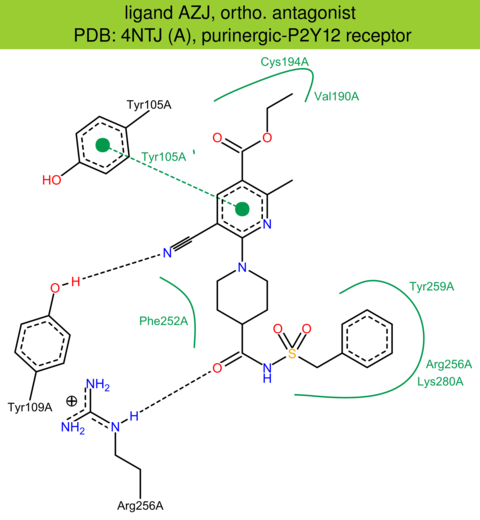 |
ortho. antagonist |
| 24 | 4O9R (A) | Q99835 | human | smoothened | 3.20 | 335/787 | cytochrome | U. Weierstall | NAT. COMMUN. | 2014 | CY8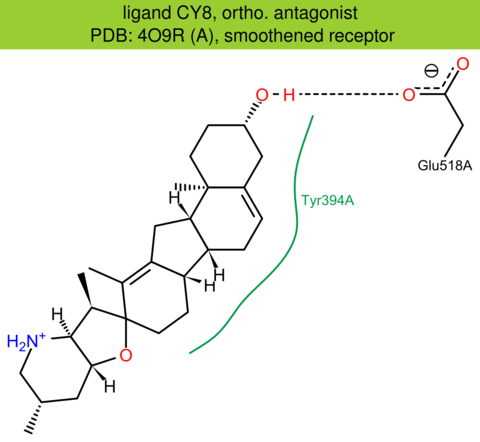 |
ortho. antagonist |
| 25 | 4OO9 (A) | P41594 | human | glutamate-R5 | 2.60 | 245/1212 | lysozyme | A.S. Dore | NATURE | 2014 | 2U8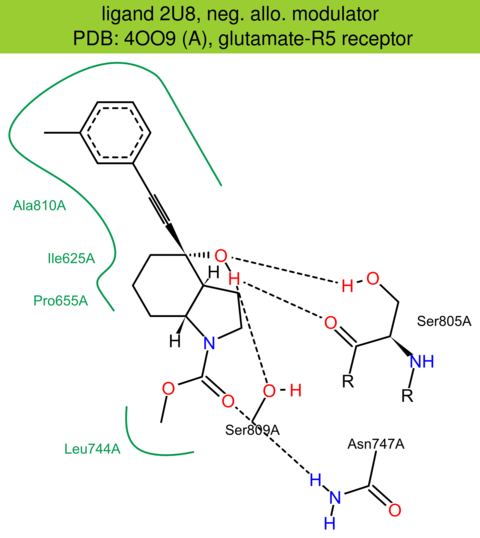 |
negative allo. modulator |
| 26 | 4OR2 (A) | Q13255 | human | glutamate-R1 | 2.80 | 255/1194 | cytochrome | H. Wu | SCIENCE | 2014 | FM9 |
negative allo. modulator |
| 27 | 4QIM (A) | Q99835 | human | smoothened | 2.61 | 351/787 | cytochrome | C. Wang | NAT. COMMUN. | 2014 | A8T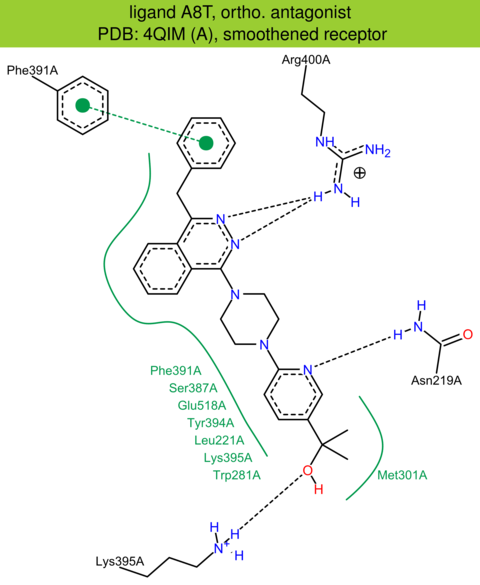 |
ortho. antagonist |
| 28 | 4RWA (A) | P41143 | human | opioid-delta | 3.28 | 286/372 | cytochrome | G. Fenalti | NAT. STRUCT. MOL. BIOL. | 2015 | peptide | ortho. antagonist |
| 29 | 4RWD (A) | P41143 | human | opioid-delta | 2.70 | 295/372 | cytochrome | G. Fenalti | NAT. STRUCT. MOL. BIOL. | 2015 | peptide |
ortho. antagonist |
| 30 | 4RWS (A) | P61073 | human | chemokine-CXCR4 | 3.10 | 275/352 | lysozyme | L. Qin | SCIENCE | 2015 | protein |
ortho. antagonist |
| 31 | 4XNV (A) | P47900 | human | purinergic-P2Y1 | 2.20 | 290/373 | ruberodoxin | D. Zhang | NATURE | 2015 | BUR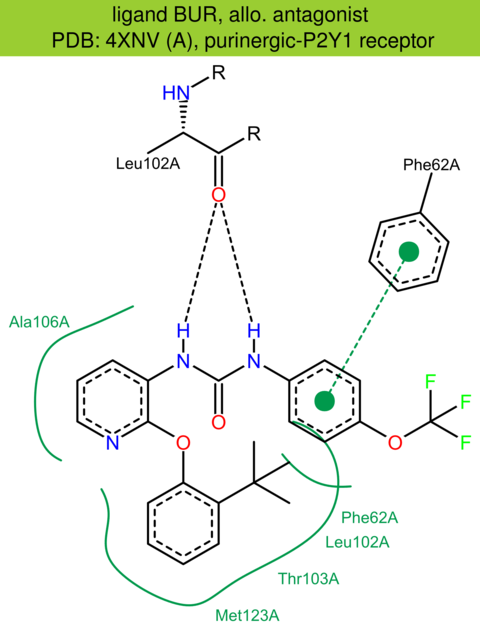 |
allo. antagonist |
| 32 | 4XNW (A) | P47900 | human | purinergic-P2Y1 | 2.70 | 292/373 | ruberodoxin | D. Zhang | NATURE | 2015 | 2ID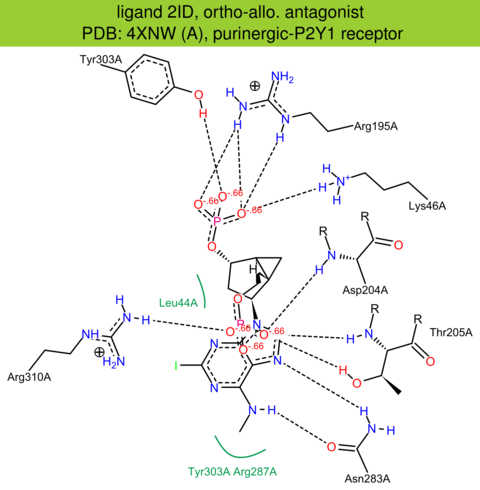 |
ortho-allo. antagonist |
| 33 | 4YAY (A) | P30556 | human | angiotensin-AT1R | 2.90 | 288/359 | cytochrome | H. Zhang | CELL | 2015 | ZD7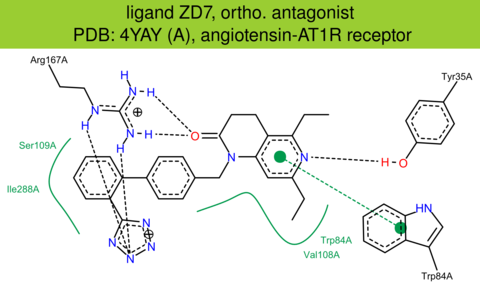 |
ortho. antagonist |
| 34 | 4Z34 (A) | Q92633 | human | lysophosphatidic-acid-R1 | 3.00 | 292/364 | cytochrome | J.E. Chrencik | CELL | 2015 | ON7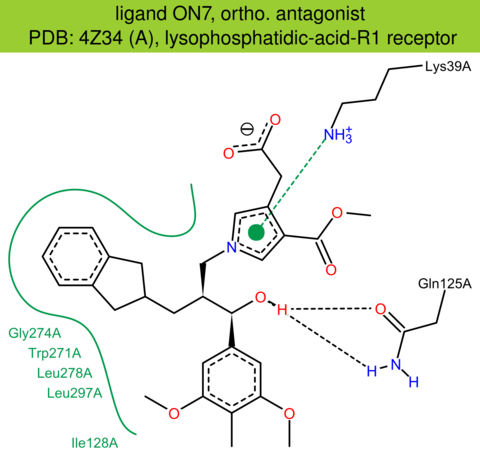 |
ortho. antagonist |
| 35 | 4Z35 (A) | Q92633 | human | lysophosphatidic-acid-R1 | 2.90 | 289/364 | cytochrome | J.E. Chrencik | CELL | 2015 | ON9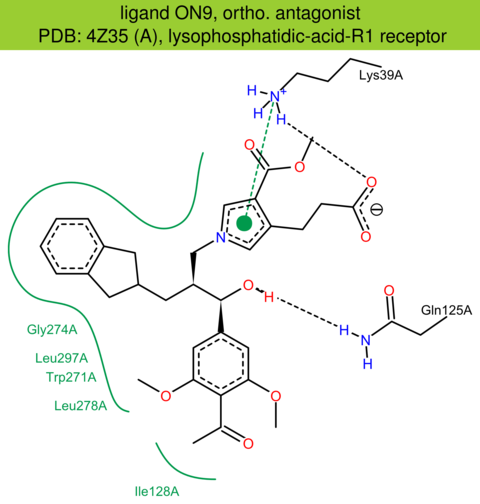 |
ortho. antagonist |
| 36 | 4Z36 (A) | Q92633 | human | lysophosphatidic-acid-R1 | 2.90 | 289/364 | cytochrome | J.E. Chrencik | CELL | 2015 | ON3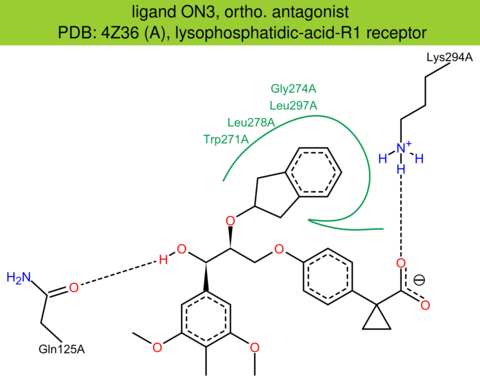 |
ortho. antagonist |
| 37 | 4Z9G (A) | P34998 | human | corticotropin-R1 | 3.18 | 260/444 | lysozyme | A.S. Dore | CURR. MOL. PHARMACOL. | 2017 | 1Q5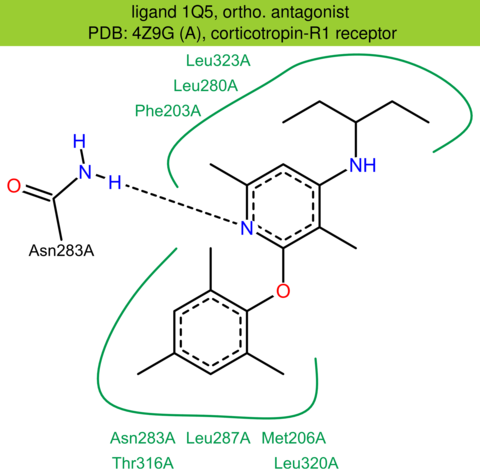 |
ortho. antagonist |
| 38 | 4ZUD (A) | P30556 | human | angiotensin-AT1R | 2.80 | 271/359 | cytochrome | H. Zhang | J. BIOL. CHEM. | 2015 | OLM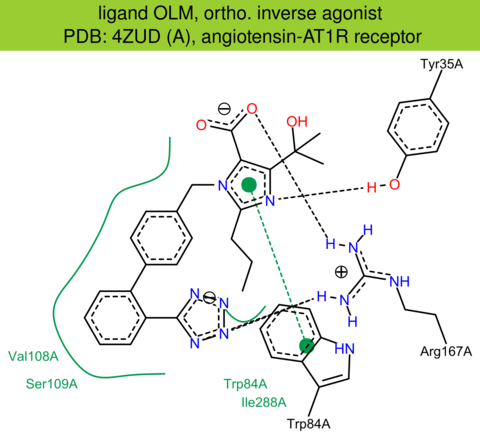 |
ortho. inverse agonist |
| 39 | 5CGC (A) | P41594 | human | glutamate-R5 | 3.10 | 248/1212 | lysozyme | J.A. Christopher | J. MED. CHEM. | 2015 | 51D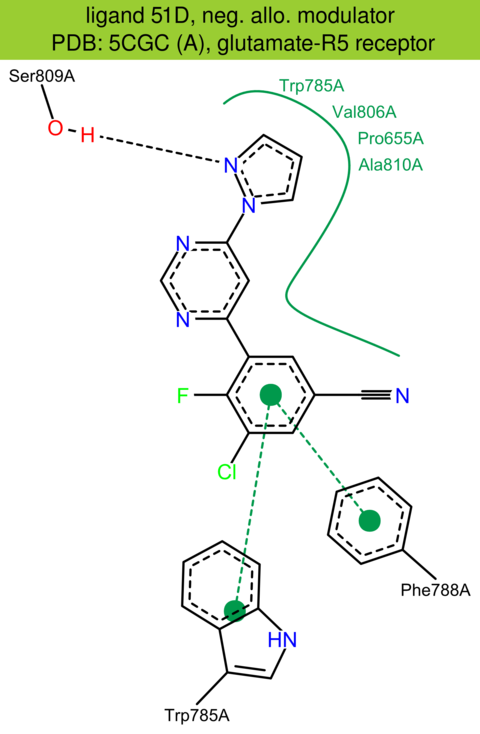 |
negative allo. modulator |
| 40 | 5CGD (A) | P41594 | human | glutamate-R5 | 2.60 | 247/1212 | lysozyme | J.A. Christopher | J. MED. CHEM. | 2015 | 51E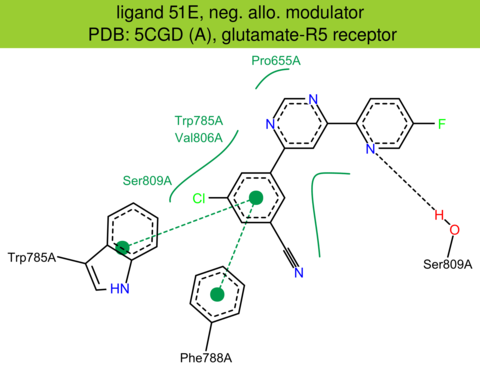 |
negative allo. modulator |
| 41 | 5D6L (A) | P07550 | human | adrenergic-beta2 | 3.20 | 280/413 | lysozyme | P. Ma | NAT. PROTOC. | 2017 | CAU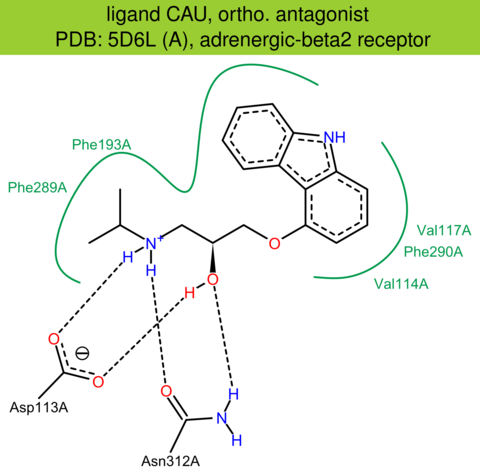 |
ortho. antagonist |
| 42 | 5DHG (A) | P41146 | human | opioid-nociceptin | 3.00 | 281/370 | cytochrome | R.L. Miller | STRUCTURE | 2015 | DGV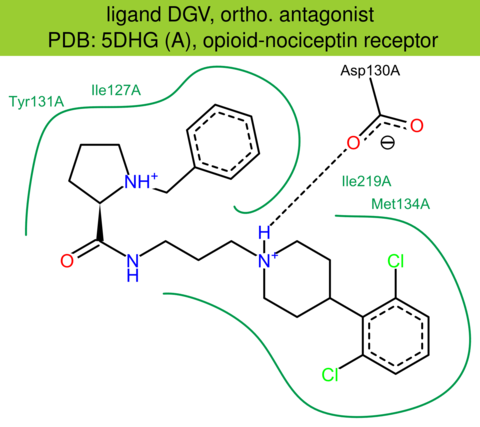 |
ortho. antagonist |
| 43 | 5DHH (A) | P41146 | human | opioid-nociceptin | 3.00 | 282/370 | cytochrome | R.L. Miller | STRUCTURE | 2015 | DGW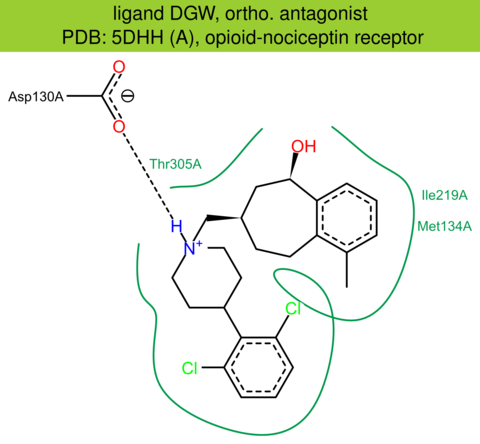 |
ortho. antagonist |
| 44 | 5EE7 (A) | P47871 | human | glucagon | 2.50 | 257/477 | lysozyme | A. Jazayeri | NATURE | 2016 | 5MV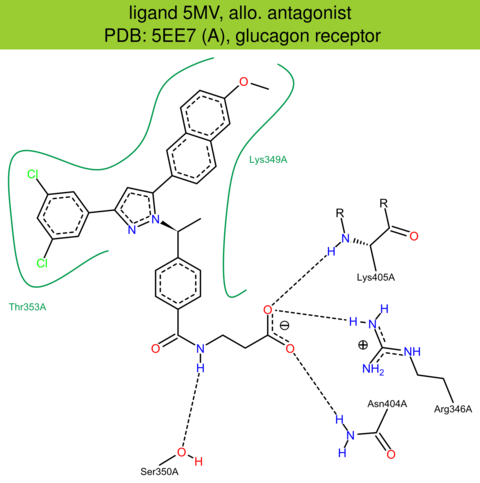 |
allo. antagonist |
| 45 | 5F8U (A) | P07700 | turkey | adrenergic-beta1 | 3.35 | 276/483 | none | A.G.W. Leslie | NAT. STRUCT. MOL. BIOL. | 2015 | P32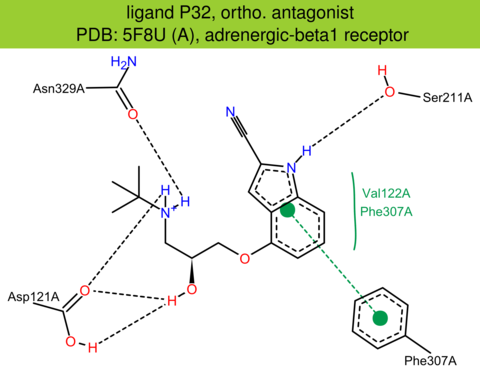 |
ortho. antagonist |
| 46 | 5GLI (A) | P24530 | human | endothelin-B | 2.50 | 312/442 | none | W. Shihoya | NATURE | 2016 | none | none |
| 47 | 5IU4 (A) | P29274 | human | adenosine-A2A | 1.72 | 294/412 | cytochrome | E. Segala | J. MED. CHEM. | 2016 | ZMA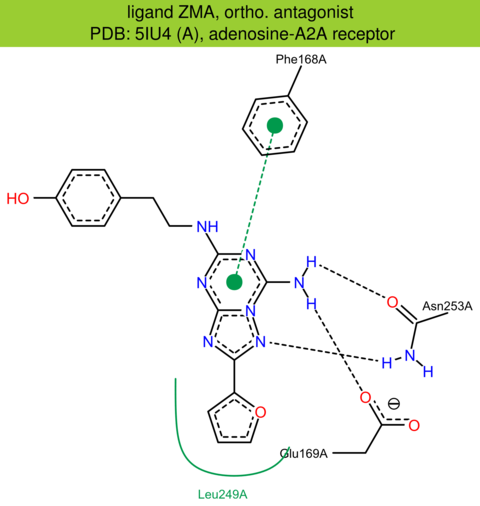 |
ortho. antagonist |
| 48 | 5JQH (A) | P00720 | human | adrenergic-beta2 | 3.20 | 283/413 | lysozyme | D.P. Staus | NATURE | 2016 | CAU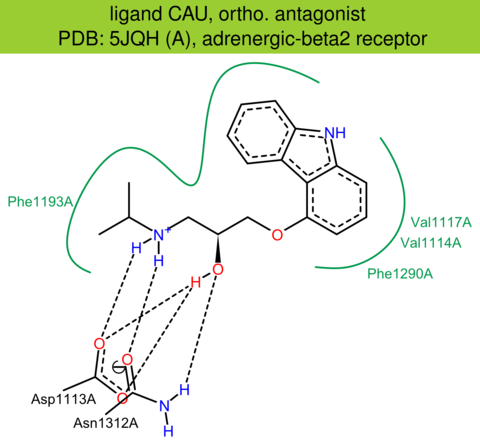 |
ortho. antagonist |
| 49 | 5L7D (A) | Q99835 | human | smoothened | 3.20 | 483/787 | cytochrome | E.F. Byrne | NATURE | 2016 | CLR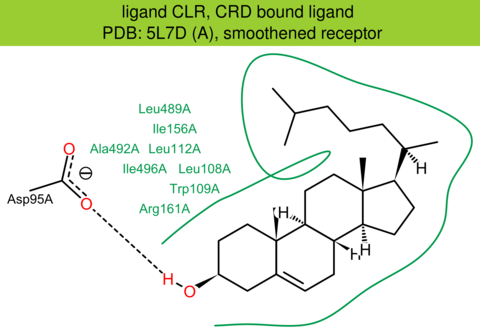 |
CRD bound ligand |
| 50 | 5L7I (A) | Q99835 | human | smoothened | 3.30 | 467/787 | cytochrome | E.F. Byrne | NATURE | 2016 | VIS |
ortho. antagonist |
| 51 | 5LWE (A) | P51686 | human | chemokine-CCR9 | 2.80 | 271/369 | none | C. Oswald | NATURE | 2016 | 79K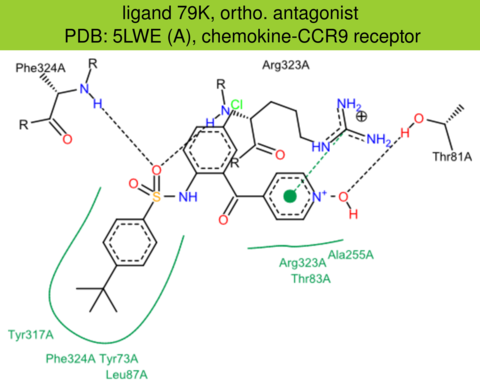 |
ortho. antagonist |
| 52 | 5NDD (A) | D9IEF7 | human | protease-activated-R2 | 2.80 | 295/397 | cytochrome | R.K.Y. Cheng | NATURE | 2017 | 8TZ |
ortho. antagonist |
| 53 | 5T1A (A) | P41597 | human | chemokine-CCR2 | 2.81 | 283/374 | lysozyme | Y. Zheng | NATURE | 2016 | VT5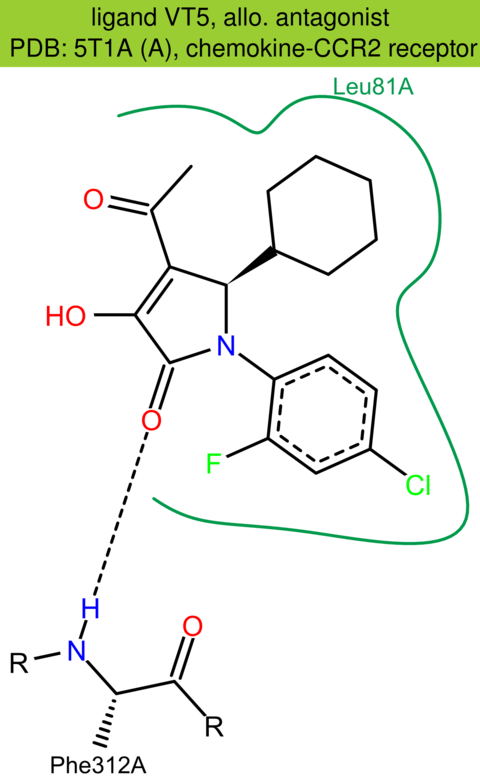 |
allo. antagonist |
| 54 | 5T1A (A) | P41597 | human | chemokine-CCR2 | 2.81 | 283/374 | lysozyme | Y. Zheng | NATURE | 2016 | 73R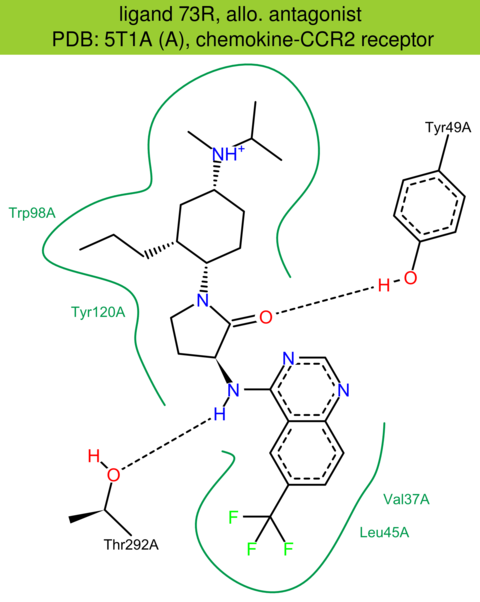 |
ortho. antagonist |
| 55 | 5TGZ (A) | P21554 | human | cannabinoid-CB1 | 2.80 | 289/472 | flavodoxin | T. Hua | CELL | 2016 | ZDG |
ortho. antagonist |
| 56 | 5U09 (A) | P21554 | human | cannabinoid-CB1 | 2.60 | 282/472 | none | Z. Shao | NATURE | 2016 | 7DY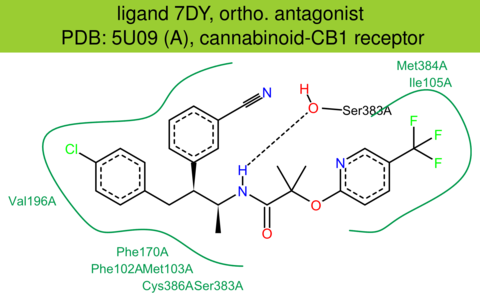 |
ortho. antagonist |
| 57 | 5UEN (A) | P30542 | human | adenosine-A1A | 3.20 | 292/326 | cytochrome | A. Glukhova | CELL | 2017 | none | none |
| 58 | 5UIG (A) | P29274 | human | adenosine-A2A | 3.50 | 288/412 | cytochrome | B. Sun | PROC. NATL. ACAD. SCI. | 2017 | 8D1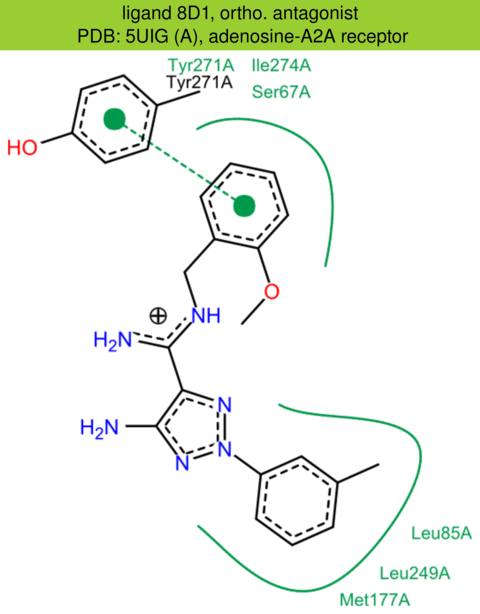 |
ortho. antagonist |
| 59 | 5VEW (A) | P43220 | human | glucagon-like-R1 | 2.70 | 262/463 | lysozyme | G. Song | NATURE | 2017 | 97Y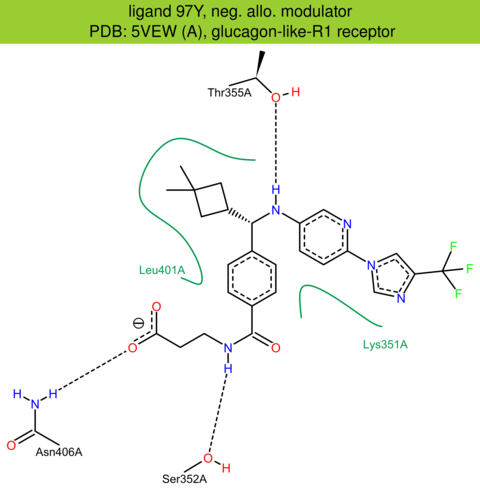 |
negative allo. modulator |
| 60 | 5VEX (A) | P43220 | human | glucagon-like-R1 | 3.00 | 262/463 | lysozyme | G. Song | NATURE | 2017 | 97V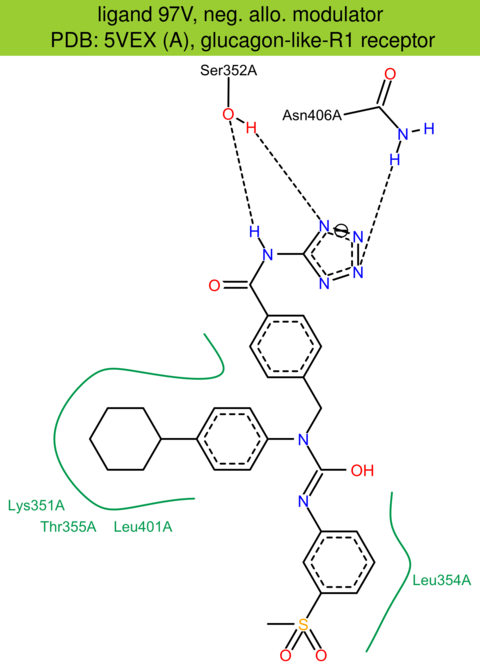 |
negative allo. modulator |
| 61 | 5XEZ (A) | P47871 | human | glucagon | 3.00 | 389/477 | lysozyme | H. Zhang | NATURE | 2017 | 97V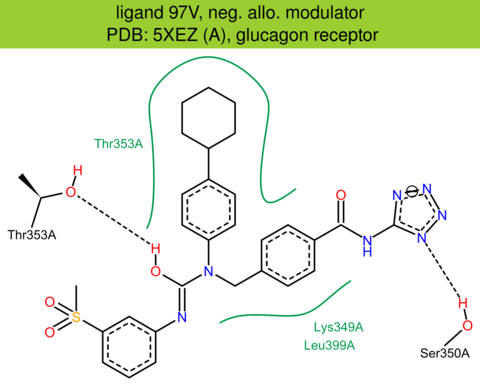 |
negative allo. modulator |
| 62 | 5XF1 (A) | P47871 | human | glucagon | 3.19 | 389/477 | lysozyme | H. Zhang | NATURE | 2017 | 97V |
negative allo. modulator |
| 63 | 6BQH (A) | P28335 | human | serotonin-2C | 2.70 | 287/458 | cytochrome | Y. Peng | CELL | 2018 | E2J |
ortho. antagonist |
Abbreviations
| ortho. | - | orthosteric |
| allo. | - | allosteric |
| neg. | - | negative |
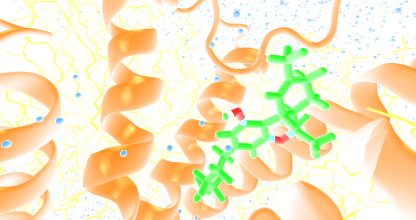
Ligand images
The images of ligand interactions were created using the PoseView service of the Center for Bioinformatics Hamburg. The details how the interactions are determined are described in [K. Stierand, M. Rarey (2010), Drawing the PDB: Protein-ligand complexes in two dimensions, ACS Med. Chem. Lett., DOI: 10.1021/ml100164p].